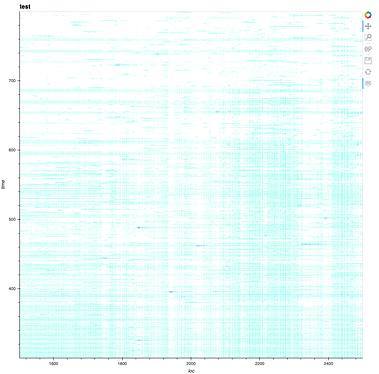
Hi,
sure!
I actually just want the plot to look exactly like the one I am producing now and attached to my previous post.
So far I have:
import datashader
from holoviews.plotting.util import process_cmap
import numpy as np
import pandas as pd
import holoviews as hv
import holoviews.operation.datashader as hd
from holoviews import dim
hv.extension('bokeh')
data2 = [
[10.0,np.array([5.0,7.0,9.0],dtype='f'),np.array([10.0,200.0,30.0],dtype='f')],
[15.0,np.array([5.2,5.6,9.7],dtype='f'),np.array([100.0,20.0,30.0],dtype='f')],
[25.0,np.array([1.1,7.2,9.3],dtype='f'),np.array([110.0,20.0,33.3],dtype='f')],
[110.0,np.array([5.0,7.0,9.0],dtype='f'),np.array([10.0,200.0,30.0],dtype='f')],
[115.0,np.array([5.2,5.6,9.7],dtype='f'),np.array([100.0,20.0,30.0],dtype='f')],
[125.0,np.array([1.1,7.2,9.3],dtype='f'),np.array([110.0,20.0,33.3],dtype='f')],
[210.0,np.array([5.0,7.0,9.0],dtype='f'),np.array([10.0,200.0,30.0],dtype='f')],
[215.0,np.array([5.2,5.6,9.7],dtype='f'),np.array([100.0,20.0,30.0],dtype='f')],
[225.0,np.array([1.1,7.2,9.3],dtype='f'),np.array([110.0,20.0,33.3],dtype='f')],
]
flatdata = np.fromiter(((series[0],point[0],point[1]) for series in data2 for point in zip(series[1],series[2])), count=27,dtype=[('time', 'f'), ('loc', 'f'), ('inty', 'f')])
df = pd.DataFrame(data=flatdata, columns=['time','loc','inty'])
points = hv.Points(df, kdims=['time', 'loc'], vdims=['inty']).opts(
fontsize={'title': 16, 'labels': 14, 'xticks': 6, 'yticks': 12},
color=np.log(dim('inty')),
colorbar=True,
cmap='Magma',
width=1000,
height=1000,
tools=['hover'])
raster = hd.rasterize(points,cmap=process_cmap("blues", provider="bokeh"),aggregator=datashader.sum('inty'),cnorm='log',alpha=50, min_alpha=10).opts(
tools=['hover']).opts(
plot = dict(
width=1000,
height=1000)
)
hv.renderer('bokeh')
hd.dynspread(raster)
The only thing I want to get rid of, is the “flatdata” line, since it is rather wasteful with millions of datapoints. I want to directly plot from the discretized data.
Thanks!