Hello,
I’m trying to make a Gridspace filled with Quadmesh objects, but every time I try to display it, my runtime crashes.
I’ve tried the suggestions here (rasterizing with DataShader, storing the underlying data in a Dask or CuPy array–though I haven’t worked with Dask or CuPy before, so I might not have done this correctly), but none seemed to help. One thing that seemed to help was instead of storing the data in a hv.Dataset object and using the .to(hv.QuadMesh) method on it, storing it in a naked xarray and calling the hv.QuadMesh() function directly on that. However, this didn’t help that much, and the downside is that it doesn’t automatically generate the slider for the z dimension when I plot it.
I know that displaying this many images of this size is a bit ambitious, but perhaps it can be done with the right mix of pre-processing and GPU use? And if it can be made to work, perhaps it would be worth adding as an example in the “Large Data” tutorial linked above?
Here’s my code (intended to be run in Google Colab):
# Imports
import xarray as xr
import numpy as np
import holoviews as hv
!pip install datashader
from holoviews.operation.datashader import rasterize
def build_images_dict(x_size, y_size, z_size, n_rows, n_columns):
'''
Function to build a 2D dictionary of images that will be used as the data source for hv.GridSpace.
In the dictionary, the key is a tuple of the image identifier ('A', 1) and the value is a HoloViews Quadmesh object.
'''
# Define row and column labels
row_labels = ['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H']
column_labels = np.arange(1,13)
## Make empty dictionary and fill with Quadmesh objects
images_dict = {}
for row_num in range(0, n_rows):
for column_num in range(0, n_columns):
# Make image data
image = xr.DataArray(np.random.rand(x_size, y_size, z_size),
dims=('x', 'y', 'z'),
name='Intensity',
coords={'x':np.arange(x_size),
'y':np.arange(y_size),
'z':np.arange(z_size)})
# Save image as HoloViews Dataset object
image_array = hv.Dataset(image)
# Make Quadmesh object from the Dataset
qm = image_array.to(hv.QuadMesh, kdims=['x', 'y'])
# Rasterize to make image size more manageable
# qm = rasterize(qm, precompute=False)
# Add Quadmesh object to dictionary of Quadmesh objects
images_dict[column_labels[column_num], row_labels[row_num]] = qm
return images_dict
# Call build_images_dict
images_dict = build_images_dict(x_size=10, y_size=10, z_size=10, n_rows=4, n_columns=4) # small sample data so code will run
# images_dict = build_images_dict(x_size=1000, y_size=1000, z_size=10, n_rows=8, n_columns=12) # this is the size of my actual data
# Make a GridSpace from the output of build_images_dict
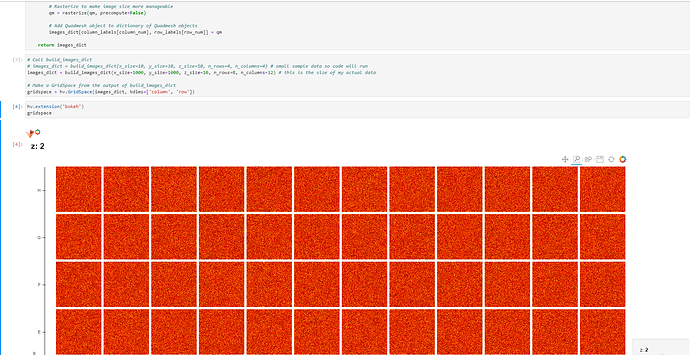
gridspace = hv.GridSpace(images_dict, kdims=['column', 'row'])
hv.extension('bokeh')
gridspace
Thanks in advance for any help!