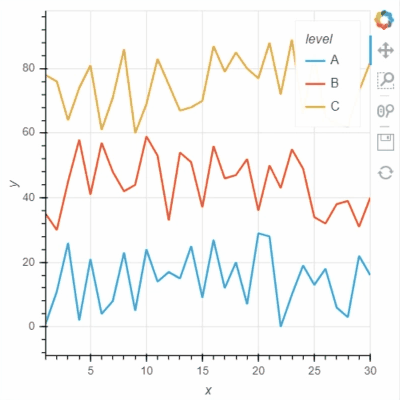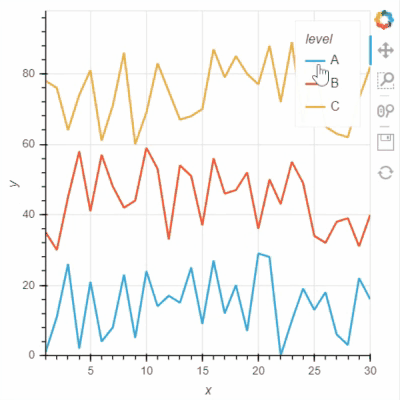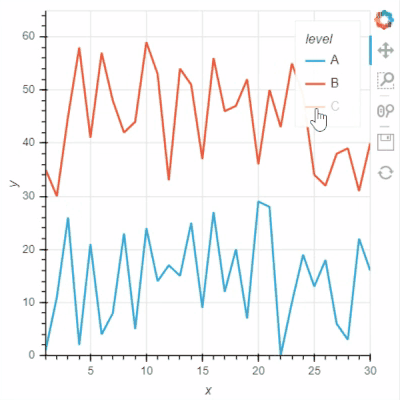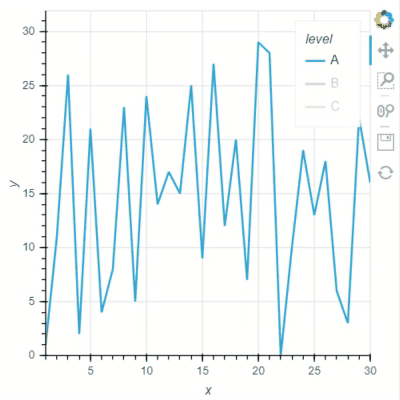
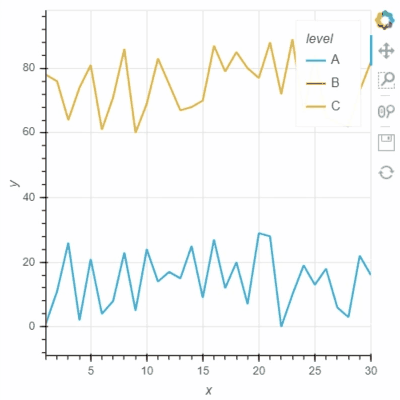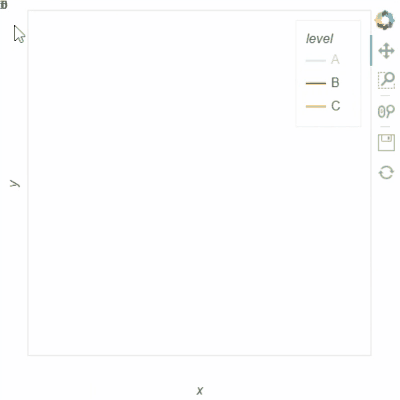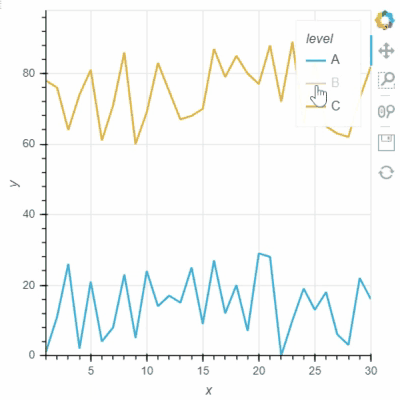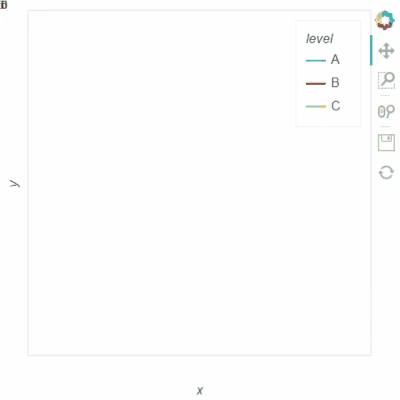
I think Bokeh has two options for “legend.click_policy”, “hide” and “mute”. If it is “hide”, the hided data points will not be selected. Not sure if Holoviews and GeoViews have a similar option for this?
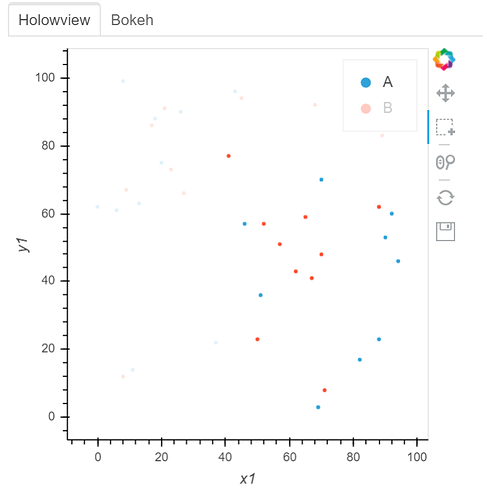
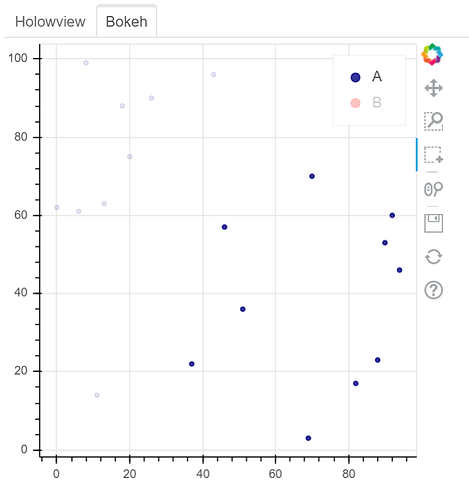
See two screenshots below. Legends “B” are muted in both two plots, and only data points of “A” are intended to be selected using Box Select tool. In Holoviews plot, the red points from Legend “B” are selected, while only Legend “A” points are selected in Bokeh plot:
Here is my example code:
data_plot_opts = dict(muted_alpha = 0.,
default_tools=[‘pan’,‘box_select’,‘reset’,‘wheel_zoom’, ‘save’],
active_tools =[‘box_select’])
y1=random.sample(range(100), 20)
x1=random.sample(range(100), 20)
x2=random.sample(range(100), 20)
y2=random.sample(range(100), 20)
Holoview:
df_trial1=pd.DataFrame({‘y1’:y1, ‘x1’:x1})
df_trial2=pd.DataFrame({‘y2’:y2, ‘x2’:x2})
data_profile_1 = hv.Points(df_trial1, kdims=[‘x1’, ‘y1’], label=“A”).opts(**data_plot_opts)
data_profile_2 = hv.Points(df_trial2, kdims=[‘x2’, ‘y2’], label=“B”).opts(**data_plot_opts)
data_profile_Holoview = pn.Column((data_profile_1*data_profile_2).opts(width=400, height=400), sizing_mode=‘stretch_width’)
Bokeh:
p = figure(plot_width=400, plot_height=400)
p.circle(x1, y1, color=“navy”, alpha=0.8, legend_label=‘A’)
p.circle(x2, y2, color=“red”, alpha=0.8, legend_label=‘B’)
p.add_tools(BoxSelectTool())
p.legend.location = “top_right”
p.legend.click_policy=“hide”
data_profile_tab = pn.Tabs((‘Holowview’, data_profile_Holoview), (‘Bokeh’, p), tabs_location=‘above’)
all_sections_view = pn.Row(data_profile_tab, width_policy=‘max’, height_policy=‘max’)
layout = all_sections_view.get_root()
curdoc().add_root(layout)
curdoc().title = “Legend_Mute”