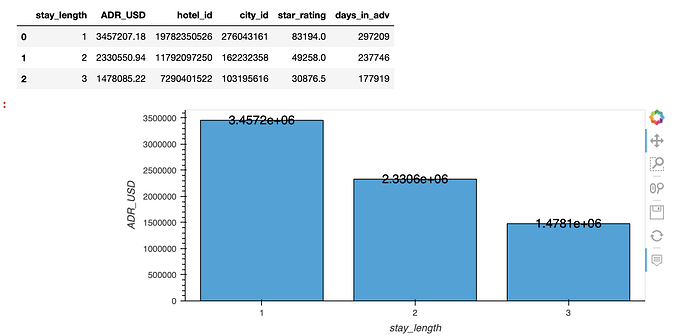
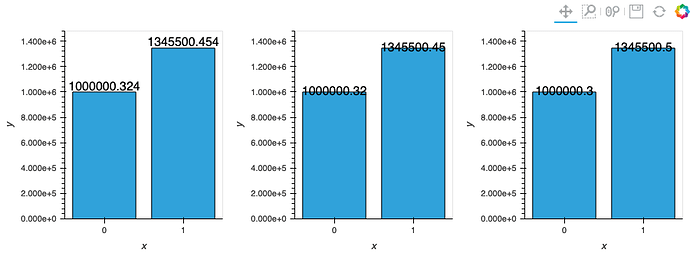
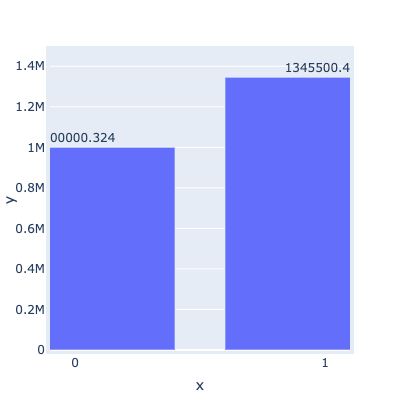
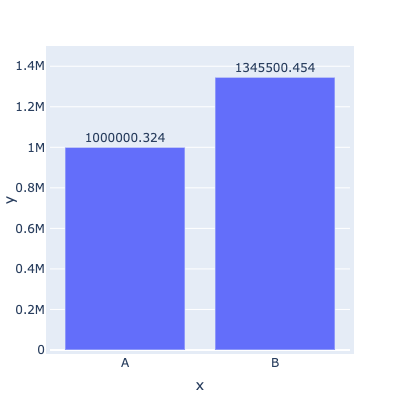
This works!!
thank you so much for your guidance on the scientific notation :))) you are awesome!
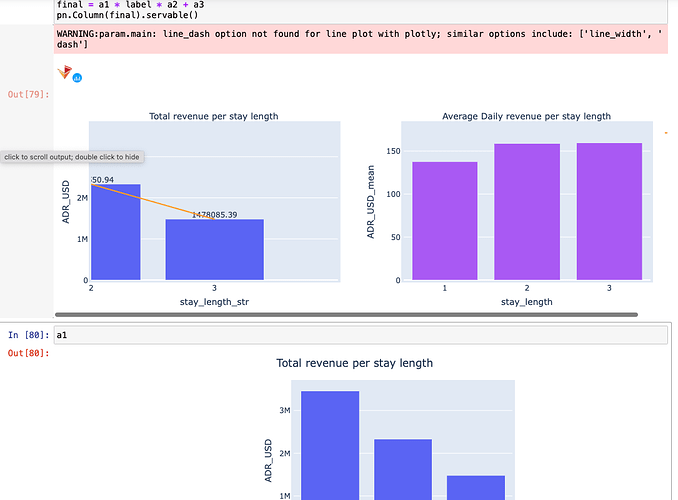
but unfortunately overlaying (with the * operator) makes the graph super duper wonky so I will just avoid Plotly all together (string column as suggested will make Labels & line plot become truncated unlike the bar plot which Plotly will align to)! (see ss)
just posting my code here for any1 who wants to reference:
# whole_df is just a pd.read_csv('on some file i have :(')
# some preprocessing of my df before plotting
whole_df.loc[:, 'ADR_USD'] = whole_df.ADR_USD.apply('{0:.2f}'.format).astype(float)
whole_df.loc[:, 'ADR_USD_mean'] = whole_df['ADR_USD'] # this is only for the formation of avriable df
df = whole_df.sort_values(["stay_length", "ADR_USD"], ascending=False).groupby(
"stay_length").agg({'ADR_USD': 'sum', 'ADR_USD_mean': 'mean'}).reset_index()
df.loc[:, 'ADR_USD_text'] = (df.ADR_USD + 150000)
df.loc[:, 'ADR_USD_text1'] = (df.ADR_USD_mean + 5)
df.loc[:, 'stay_length_str'] = df.stay_length.astype(str)
# barplot
a1 = df.hvplot.bar(
x='stay_length_str', y="ADR_USD", title='Total revenue per stay length'
).opts(height=350, width=450,
)
# line plot
a2 = df.hvplot.line(
x='stay_length_str', y="ADR_USD",
line_dash='dashed', # different parameters for different extensions, sigh, expected
# dash='dashdot',
).opts(height=350, width=450, padding =0.2, color='orange'
)
# another bar plot
a3 = df.hvplot.bar(x='stay_length', y="ADR_USD_mean", title='Average Daily revenue per stay length',
color='orange'
).opts(height=350, width=450, padding=1
)
# labels
df['labels'] = (df.ADR_USD).astype(str)
# value_dimension = hv.Dimension('ADR_USD', value_format=lambda x: '%.1f' % x)
label = hv.Labels(df, ['stay_length', 'ADR_USD'],
# value_dimension,
'labels',
).opts(ylim=(0,1.1*df.ADR_USD.max()), yoffset=100000)
# display
hv.extension('bokeh')
final = a1 * label * a2 + a3
# ls(final)
pn.Column(final).servable()
Also just to note using ls() = hv.link_selections.instance() will lead to no attribute error: ‘Labels’ object has no attribute ‘_empty_region’.