Marc
April 28, 2020, 2:07pm
1
THIS QUESTION AND ANSWER IS COPIED FROM https://github.com/holoviz/panel/issues/1307 .
I’d like to incorporate a protein viewer in my panel application. To do so I’m looking at ngl which has both an ipywidget for notebooks and JS/HTML available for embedding.
How do I do this?
Marc
April 28, 2020, 2:07pm
2
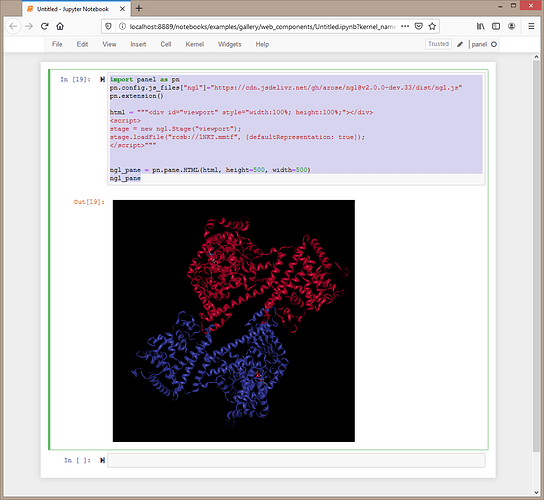
This code will work
import panel as pn
pn.config.js_files["ngl"]="https://cdn.jsdelivr.net/gh/arose/ngl@v2.0.0-dev.33/dist/ngl.js"
pn.extension()
html = """<div id="viewport" style="width:100%; height:100%;"></div>
<script>
stage = new ngl.Stage("viewport");
stage.loadFile("rcsb://1NKT.mmtf", {defaultRepresentation: true});
</script>"""
ngl_pane = pn.pane.HTML(html, height=500, width=500)
ngl_pane
1 Like
Jhsmit
August 25, 2020, 7:21pm
3
I was playing around with this and turned it into a ‘component’ for panel following @Marc ’s guide on subclassing the HTML pane.
The code is below, should I contribute it to awesome-panel?
Is there already a way of getting variables from the html to python? The stage object has some variables I’d like to access from python.
import panel as pn
import param
pn.config.js_files["ngl"]="https://unpkg.com/ngl@2.0.0-dev.37/dist/ngl.js"
pn.extension()
class NGLViewer(pn.pane.HTML):
pdb_string = param.String()
rcsb_id = param.String()
representation = param.Selector(default='ribbon',
objects=['ball+stick', 'backbone', 'ball+stick', 'cartoon', 'hyperball', 'licorice',
'ribbon', 'rope', 'spacefill', 'surface'])
spin = param.Boolean(default=False)
priority = 0
_rename = dict(pn.pane.HTML._rename, pdb_string=None, rcsb_id=None, representation=None, spin=None)
def __init__(self, **params):
super().__init__(**params)
self.load_string = \
f"""
stage = new NGL.Stage("viewport");
stage.loadFile()"""
self._update_object_from_parameters()
@param.depends('representation', 'spin', watch=True)
def _update_object_from_parameters(self):
html =\
f"""
<div id="viewport" style="width:100%; height:100%;"></div>
<script>
{self.load_string}.then(function(o){{
o.addRepresentation("{self.representation}");
o.autoView();
}}
);
stage.setSpin({'true' if self.spin else 'false'});
</script>
"""
self.object = html
@param.depends('pdb_string', watch=True)
def _update_object_from_pdb_string(self):
self.load_string = \
f"""
var PDBString = `{self.pdb_string}`;
stage = new NGL.Stage("viewport");
stage.loadFile( new Blob([PDBString], {{type: 'text/plain'}}), {{ ext:'pdb'}} )"""
self._update_object_from_parameters()
@param.depends('rcsb_id', watch=True)
def _update_object_from_rcsb_id(self):
self.load_string = \
f"""
stage = new NGL.Stage("viewport");
stage.loadFile("rcsb://{self.rcsb_id}")"""
self._update_object_from_parameters()
class ProteinViewer(param.Parameterized):
input_option = param.Selector(objects=['Upload File', 'RCSB PDB'])
rcsb_id = param.String()
load_structure = param.Action(lambda self: self._load_structure())
def __init__(self, **param):
super(ProteinViewer, self).__init__(**param)
self.file_widget = pn.widgets.FileInput(accept='.pdb')
self.ngl_html = NGLViewer(height=500, width=500)
def _load_structure(self):
if self.input_option == 'Upload File':
if self.file_widget.value:
string = self.file_widget.value.decode()
self.ngl_html.pdb_string = string
else:
pass
elif self.input_option == 'RCSB PDB':
self.ngl_html.rcsb_id = self.rcsb_id
def view(self):
col = pn.Column(*pn.Param(self.param))
col.insert(2, self.file_widget)
col.append(self.ngl_html.param.representation)
col.append(self.ngl_html.param.spin)
app = pn.Row(
col,
self.ngl_html
)
return app
pv = ProteinViewer()
pn.serve(pv.view())
1 Like
Marc
August 25, 2020, 11:59pm
4
I would be happy to review, maybe improve it, (temporarily?) host it in awesome-panel-extensions package.
I believe i might convert it to a bokeh extension, if it there are events we would like to Catch and data we would like to transfer back to python?
But i think @philippjfr should guide whether he would already now like to take it into Panel.