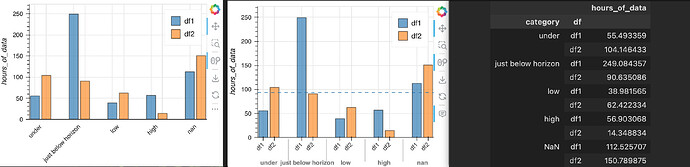
Hello!
I’m plotting a multiindex dataframe with two levels of indices, I only wanted one of them to show up and this command worked perfectly .opts(multi_level=False). However, the moment I overlayed the plot with a horizontal line using good_plot *= hv.HLine(line_height), the original multilevel x axis would come back. Is there a way around this? Here’s a toy example:
toy_csv = pd.read_csv("toy.csv", index_col=[0, 1]).rename_axis(index=["category", "df"])
plot_mean = np.mean(toy_csv['hours_of_data'])
toy_plot = toy_csv.hvplot.bar(
y="hours_of_data",
by="df",
stacked=False,
legend=False,
rot=XROTATION,
color=["#1F77B4", "#FF7F0E"],
).opts(
xlabel="",
shared_axes=False,
show_grid=True,
width=PLOT_WIDTH,
multi_level=False,
show_legend=True,
legend_position = 'top_right'
)
toy_plot
Would produce image 1. But if a horizontal line is added as an overlay plot:
toy_plot *= hv.HLine(plot_mean).opts(
color="#1F77B4",
line_dash="dashed",
line_width=1,
)
toy_plot
It would revert back to the multi-level x axis (image 2), which isn’t what I want. Any ideas on how to keep it the same? The toy example csv structure is included below as image 3 (for some reason new users can’t add attachments and can only add 1 picture? I just used random numbers)
Thank you so much!