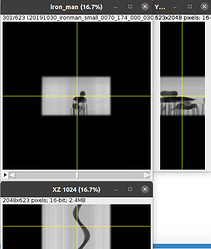
Hello @KedoKudo, this is not exactly the interaction you ask for but this can be a starting point.
# option1 need to install scikit-image and pooch
import skimage
cells = skimage.data.cells3d()[:,1,:,:]
# option2 download "https://gitlab.com/scikit-image/data/-/raw/master/cells3d.tif"
#cells = skimage.io.imread('cells3d.tif')[:,1,:,:]
import holoviews as hv
from holoviews import streams
import xarray as xr
import hvplot.xarray
import panel as pn
import numpy as np
hv.extension('bokeh')
wz, wx, wy = cells.shape
xs,ys,zs = np.arange(256), np.arange(255,-1,-1), np.arange(60)
da = xr.DataArray(cells,dims=['z','x','y'],coords=dict(z=zs,x=xs,y=ys),name='intensity')
def crosshair(x,y):
x = 0 if x is None else np.clip(x,0,255)
y = 0 if y is None else np.clip(y,0,255)
return hv.HLine(y)*hv.VLine(x)
def ximage(x,y):
x = 0 if x is None else int(np.clip(x,0,255))
return da.sel(x=x).hvplot('z','y',cmap='gray').opts(tools=[],frame_width=wz,frame_height=wy,title='')
def yimage(x,y):
y = 0 if y is None else int(np.clip(y,0,255))
return da.sel(y=y).hvplot(cmap='gray').opts(tools=[],frame_width=wx,frame_height=wz,title='')
def view(z):
img = da.sel(z=z).hvplot('x','y',cmap='gray').opts(tools=[],frame_width=wx,frame_height=wy,title='')
pointer = streams.PointerXY(x=0,y=0,source=img)
zimg = img*hv.DynamicMap(crosshair,streams = [pointer])
ximg = hv.DynamicMap(ximage,streams = [pointer])
yimg = hv.DynamicMap(yimage,streams = [pointer])
return (zimg+ximg+yimg).cols(2)
zw = pn.widgets.IntSlider(start=0,end=60,name='z')
pn.interact(view,z=zw)
![]()