I’m sure I did everything wrong: I don’t know JS good enough to dig into it, I don’t know Parameterized (and prefer pure widget solution if possible), I’m still learning Panel (by doing, instead of reading docs).
So, @Marc , thank you for your post! Inspired by it I put together this
import panel as pn
pn.extension()
import param
class SMILESViewer(param.Parameterized):
smiles = param.String(
doc="""
The value describes a molecule to be loaded into the viewer in SMILES format."""
)
opacity = param.Number(
default=0.5,
bounds=(0.0, 1.0),
step=0.01,
doc="""
Mol alpha (default: 0.5).""",
)
height = param.Number(
default=400,
bounds=None,
step=1,
doc="""
Height.""",
)
width = param.Number(
default=400,
bounds=None,
step=1,
doc="""
Width.""",
)
select = param.String(doc="""The value is an AtomSpec selection specification.""")
style = param.ObjectSelector(
default="stick",
objects=["line", "cross", "stick", "sphere", "cartoon"],
doc="""The value is a style specification. One of 'line', 'cross', 'stick', 'sphere' or 'cartoon'. Default is 'stick'""",
)
surface = param.Boolean(default=False, doc="""A surface style specification""")
zoomto = param.Boolean(default=True, doc="""An AtomSpec selection specification to zoom to""")
def __init__(self, smiles=None, surface=False, style="stick", height=400, width=400, zoomto=True, opacity=0.5, **params):
super().__init__(**params)
self.view = pn.pane.HTML(height=height, width=width)
self.height = height
self.width = width
self.smiles = smiles
self.style = style
self.surface = surface
self.opacity = opacity
self.zoomto = zoomto
def _get_html_(self):
from rdkit import Chem
import py3Dmol
assert self.style in ('line', 'stick', 'sphere', 'carton')
#smi = 'COc3nc(OCc2ccc(C#N)c(c1ccc(C(=O)O)cc1)c2P(=O)(O)O)ccc3C[NH2+]CC(I)NC(=O)C(F)(Cl)Br'
smi = self.smiles
mol = self._smi2conf(smi)
mblock = Chem.MolToMolBlock(mol)
viewer = py3Dmol.view(width=self.width, height=self.height)
viewer.addModel(mblock, 'mol')
viewer.setStyle({self.style:{}})
if self.surface:
viewer.addSurface(py3Dmol.SAS, {'opacity': self.opacity})
if self.zoomto:
viewer.zoomTo()
return viewer
@param.depends(
"smiles",
"opacity",
"height",
"width",
"style",
"surface",
"zoomto",
watch=True,
)
def _update_view(self, *events):
self.view.object = self._get_html_()
def _repr_html(self):
return self.view.object
def _smi2conf(self,smiles):
'''Convert SMILES to rdkit.Mol with 3D coordinates'''
from rdkit import Chem
from rdkit.Chem import AllChem
mol = Chem.MolFromSmiles(self.smiles)
if mol is not None:
mol = Chem.AddHs(mol)
AllChem.EmbedMolecule(mol)
AllChem.MMFFOptimizeMolecule(mol, maxIters=200)
return mol
else:
return None
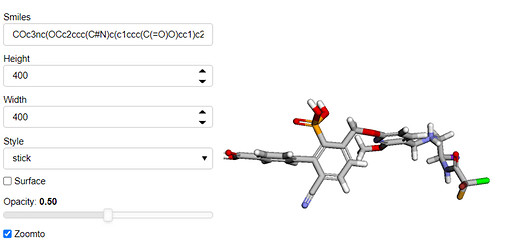
jsmol = SMILESViewer(smiles='COc3nc(OCc2ccc(C#N)c(c1ccc(C(=O)O)cc1)c2P(=O)(O)O)ccc3C[NH2+]CC(I)NC(=O)C(F)(Cl)Br')
jsmol.view.object
Which works.
But what I really want is for the last line to be jsmol.view.
When I do it - I get this error:
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
/opt/lib/python3.8/site-packages/IPython/core/formatters.py in __call__(self, obj, include, exclude)
968
969 if method is not None:
--> 970 return method(include=include, exclude=exclude)
971 return None
972 else:
/opt/lib/python3.8/site-packages/panel/viewable.py in _repr_mimebundle_(self, include, exclude)
586 doc = _Document()
587 comm = state._comm_manager.get_server_comm()
--> 588 model = self._render_model(doc, comm)
589 ref = model.ref['id']
590 manager = CommManager(comm_id=comm.id, plot_id=ref)
/opt/lib/python3.8/site-packages/panel/viewable.py in _render_model(self, doc, comm)
425 if comm is None:
426 comm = state._comm_manager.get_server_comm()
--> 427 model = self.get_root(doc, comm)
428
429 if config.embed:
/opt/lib/python3.8/site-packages/panel/pane/base.py in get_root(self, doc, comm, preprocess)
252 doc = init_doc(doc)
253 if self._updates:
--> 254 root = self._get_model(doc, comm=comm)
255 else:
256 root = self.layout._get_model(doc, comm=comm)
/opt/lib/python3.8/site-packages/panel/pane/markup.py in _get_model(self, doc, root, parent, comm)
39
40 def _get_model(self, doc, root=None, parent=None, comm=None):
---> 41 model = self._bokeh_model(**self._get_properties())
42 if root is None:
43 root = model
/opt/lib/python3.8/site-packages/panel/pane/markup.py in _get_properties(self)
77 if hasattr(text, '_repr_html_'):
78 text = text._repr_html_()
---> 79 return dict(properties, text=escape(text))
80
81
/opt/lib/python3.8/html/__init__.py in escape(s, quote)
17 translated.
18 """
---> 19 s = s.replace("&", "&") # Must be done first!
20 s = s.replace("<", "<")
21 s = s.replace(">", ">")
AttributeError: 'NoneType' object has no attribute 'replace'
With this in the output cell:
HTML(view, height=400, sizing_mode='fixed', width=400)
One more annoying thing is that when I do use jsmol.view.object I get this <py3Dmol.view at 0x2b8c63aa1ac0> following the image. I wonder if that can be removed.
P.S. As you noticed I’m interested in visualizing by SMILES notation and not from file. There is a reason for that which originates from the researcher I’m working with.
Would be really appreciated for any advise!
Thanks in Advance!
 ) illustrate few famous molecules and animate how protein folds would be a good marketing idea.
) illustrate few famous molecules and animate how protein folds would be a good marketing idea.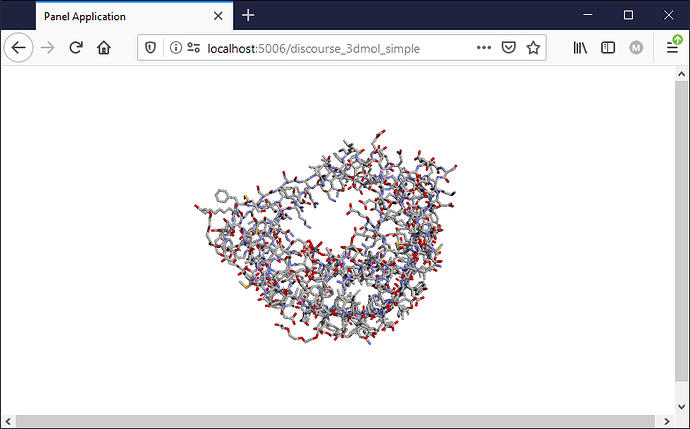

 Thanks.
Thanks.