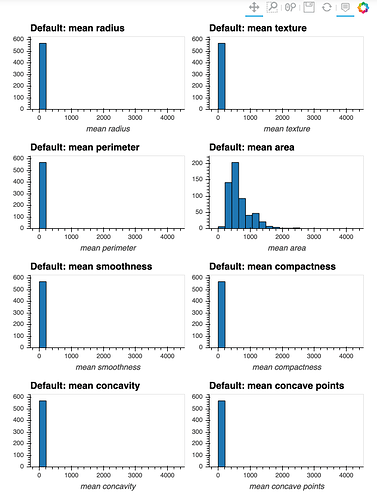
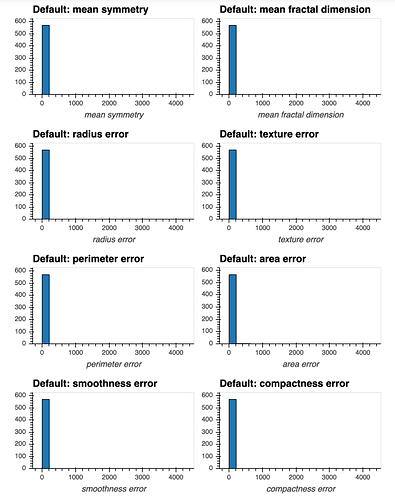
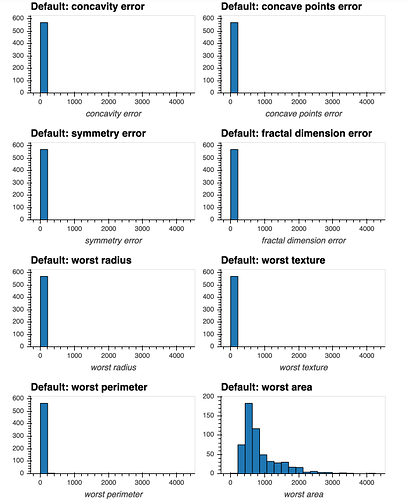
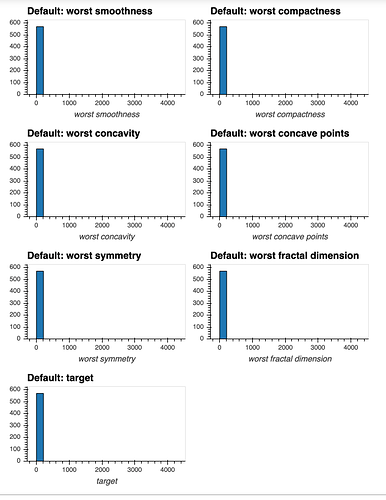
I have a dataset that contains many numeric attributes of different distributions.
In [1]: import pandas as pd
...: import numpy as np
...: import holoviews as hv
...: import hvplot.pandas # noqa
...: import feather
...: hv.extension("bokeh","plotly")
In [2]: df = pd.read_feather("../df_discourse_question1.feather")
In [3]: df
Out[3]:
mean radius mean texture mean perimeter mean area mean smoothness mean compactness mean concavity ... worst compactness worst concavity worst concave points worst symmetry worst fractal dimension target target_name
0 17.99 10.38 122.80 1001.0 0.11840 0.27760 0.30010 ... 0.66560 0.7119 0.2654 0.4601 0.11890 0 benign
1 20.57 17.77 132.90 1326.0 0.08474 0.07864 0.08690 ... 0.18660 0.2416 0.1860 0.2750 0.08902 0 benign
2 19.69 21.25 130.00 1203.0 0.10960 0.15990 0.19740 ... 0.42450 0.4504 0.2430 0.3613 0.08758 0 benign
3 11.42 20.38 77.58 386.1 0.14250 0.28390 0.24140 ... 0.86630 0.6869 0.2575 0.6638 0.17300 0 benign
4 20.29 14.34 135.10 1297.0 0.10030 0.13280 0.19800 ... 0.20500 0.4000 0.1625 0.2364 0.07678 0 benign
.. ... ... ... ... ... ... ... ... ... ... ... ... ... ... ...
564 21.56 22.39 142.00 1479.0 0.11100 0.11590 0.24390 ... 0.21130 0.4107 0.2216 0.2060 0.07115 0 benign
565 20.13 28.25 131.20 1261.0 0.09780 0.10340 0.14400 ... 0.19220 0.3215 0.1628 0.2572 0.06637 0 benign
566 16.60 28.08 108.30 858.1 0.08455 0.10230 0.09251 ... 0.30940 0.3403 0.1418 0.2218 0.07820 0 benign
567 20.60 29.33 140.10 1265.0 0.11780 0.27700 0.35140 ... 0.86810 0.9387 0.2650 0.4087 0.12400 0 benign
568 7.76 24.54 47.92 181.0 0.05263 0.04362 0.00000 ... 0.06444 0.0000 0.0000 0.2871 0.07039 1 malignant
[569 rows x 32 columns]
In [7]: df.describe()
Out[7]:
mean radius mean texture mean perimeter mean area mean smoothness mean compactness mean concavity ... worst smoothness worst compactness worst concavity worst concave points worst symmetry worst fractal dimension target
count 569.000000 569.000000 569.000000 569.000000 569.000000 569.000000 569.000000 ... 569.000000 569.000000 569.000000 569.000000 569.000000 569.000000 569.000000
mean 14.127292 19.289649 91.969033 654.889104 0.096360 0.104341 0.088799 ... 0.132369 0.254265 0.272188 0.114606 0.290076 0.083946 0.627417
std 3.524049 4.301036 24.298981 351.914129 0.014064 0.052813 0.079720 ... 0.022832 0.157336 0.208624 0.065732 0.061867 0.018061 0.483918
min 6.981000 9.710000 43.790000 143.500000 0.052630 0.019380 0.000000 ... 0.071170 0.027290 0.000000 0.000000 0.156500 0.055040 0.000000
25% 11.700000 16.170000 75.170000 420.300000 0.086370 0.064920 0.029560 ... 0.116600 0.147200 0.114500 0.064930 0.250400 0.071460 0.000000
50% 13.370000 18.840000 86.240000 551.100000 0.095870 0.092630 0.061540 ... 0.131300 0.211900 0.226700 0.099930 0.282200 0.080040 1.000000
75% 15.780000 21.800000 104.100000 782.700000 0.105300 0.130400 0.130700 ... 0.146000 0.339100 0.382900 0.161400 0.317900 0.092080 1.000000
max 28.110000 39.280000 188.500000 2501.000000 0.163400 0.345400 0.426800 ... 0.222600 1.058000 1.252000 0.291000 0.663800 0.207500 1.000000
[8 rows x 31 columns]
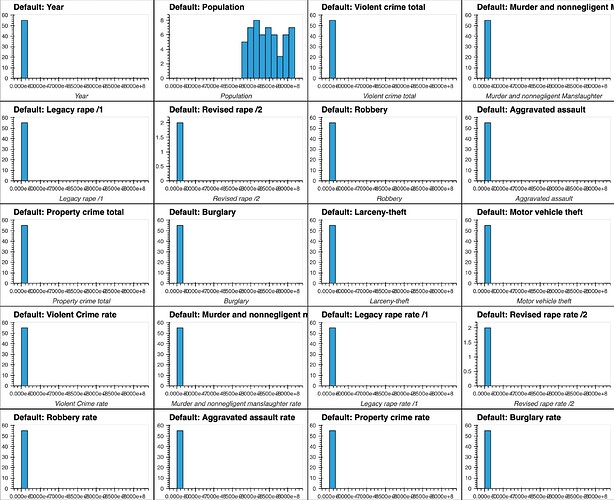
If I attempt to produce histograms for each of the numeric attributes using subplots = True and shared_axes = False , then the subplots are successfully produced, however all of the plots appear to still share an x and y axis:
In [8]: # THIS WILL PRODUCE HISTOGRAMS FOR ALL NUMERIC ATTRIBUTES IN DATAFRAME df, INCLUDING OUR "target" COLUMN:
...: df.hvplot.hist(width=300, height=200,
...: subplots=True,
...: shared_axes=False
...: ).cols(2)
Out[8]:
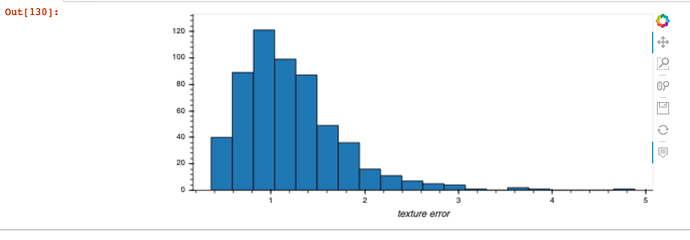
This is confirmed when I produce a single plot for one of the attributes whose histogram above was a single bin - now I can successfully see the distribution of the attribute (due to appropriate x and y axis scale being used on the plot):
In [9]: df.hvplot.hist(y="texture error")
Out[9]:
Does anyone have any idea what is happening here?
Thanks
Software Versions:
pandas 1.0.3 py37h6c726b0_0
numpy 1.18.1 py37h7241aed_0
holoviews 1.13.1 py_0 pyviz
hvplot 0.5.2 py_0 pyviz
feather-format 0.4.0 py_1003 conda-forge
bokeh 1.4.0 py37_0
plotly 4.5.4 py_0 plotly