I am just getting started this week with Holoviews (although I have used Panel to deploy an app).
N.B. I also asked about this on stackoverflow, if anyone answered there I will cross-post the answer here.
I have a number of time series in text files loaded as Pandas DataFrames where:
- each file is for a specific location
- at each location about 10 time series were collected, each with about 15,000 points
I am building a small interactive tool where a Selector can be used to choose the location / DataFrame, and then another Selector to pick 3 of 10 of the time series to be plotted together.
My goal is to allow linked zooms (both x and y scales). The questions and code will focus on this aspect of the tool.
I cannot share the actual data I am using, unfortunately, as it is proprietary, but I have created 3 random walks with specific data ranges that are consistent with the actual data.
## preliminaries ##
import pandas as pd
import numpy as np
import holoviews as hv
from holoviews.util.transform import dim
from holoviews.selection import link_selections
from holoviews import opts
from holoviews.operation.datashader import shade, rasterize
import hvplot.pandas
hv.extension('bokeh', width=100)
## create random walks (one location) ##
data_df = pd.DataFrame()
npoints=15000
np.random.seed(71)
x = np.arange(npoints)
y1 = 1300+2.5*np.random.randn(npoints).cumsum()
y2 = 1500+2*np.random.randn(npoints).cumsum()
y3 = 3+np.random.randn(npoints).cumsum()
data_df.loc[:,'x'] = x
data_df.loc[:,'rand1'] = y1
data_df.loc[:,'rand2'] = y2
data_df.loc[:,'rand3'] = y3
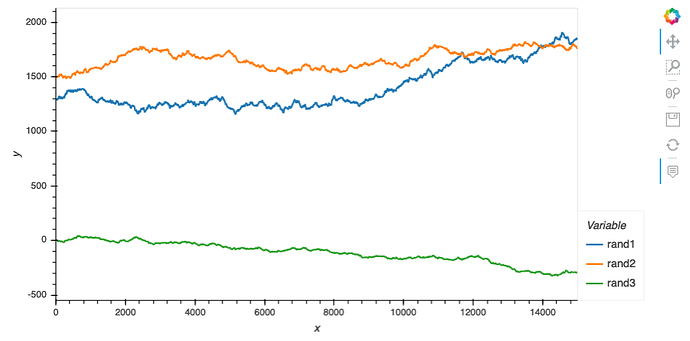
This first block is just to plot the data and show how, by design, one of the random walks have different range from the other two:
data_df.hvplot(x='x', y=['rand1', 'rand2', 'rand3'], value_label='y', width=800, height=400)
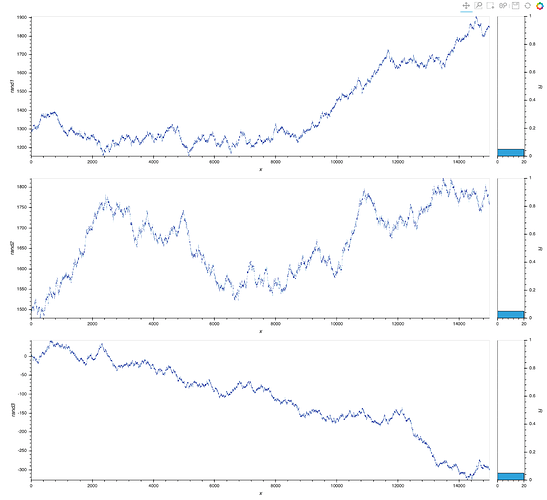
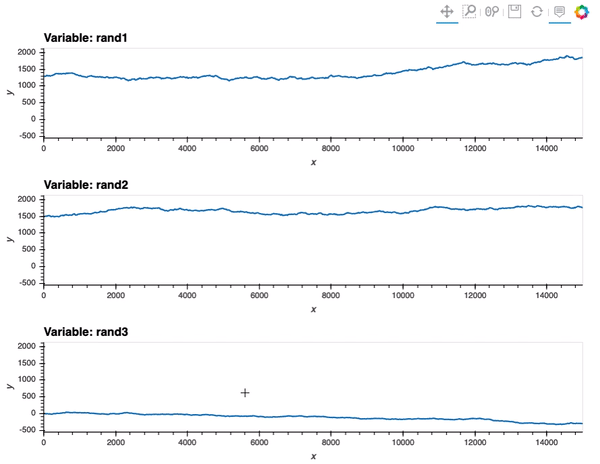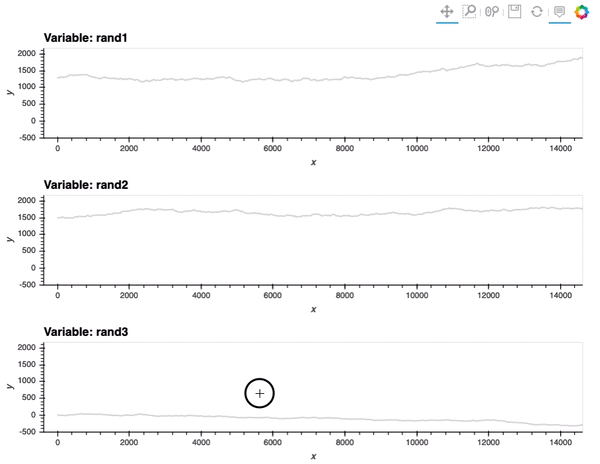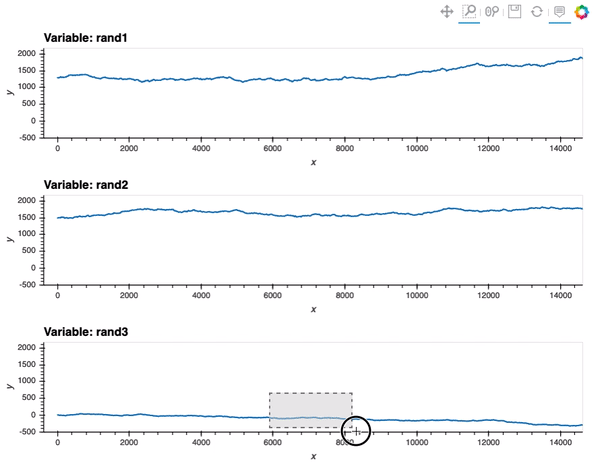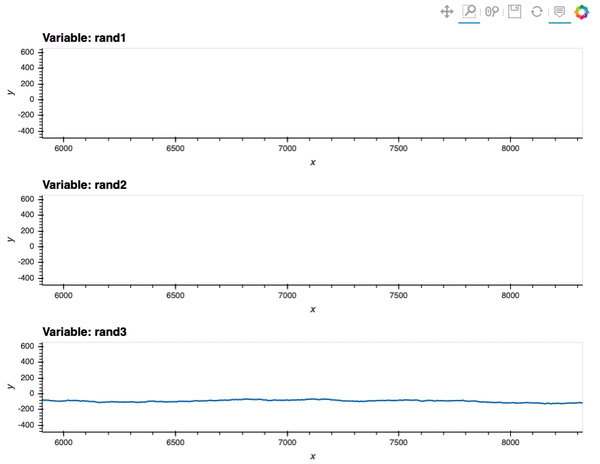
As a result, although hvplot subplots work out of the box (for linking), ranges are different so the scaling is not quite there:
data_df.hvplot(x='x', y=['rand1', 'rand2', 'rand3'], value_label='y', subplots=True, width=800, height=200).cols(1)
So, my first attempt was to adapt the Python-based Points example from Linked brushing in the documentation:
colors = hv.Cycle('Category10').values
dims = ['rand1', 'rand2', 'rand3']
layout = hv.Layout([
hv.Points(data_df, dim).opts(color=c)
for c, dim in zip(colors, [['x', d] for d in dims])
])
link_selections(layout).opts(opts.Points(width=1200, height=300)).cols(1)
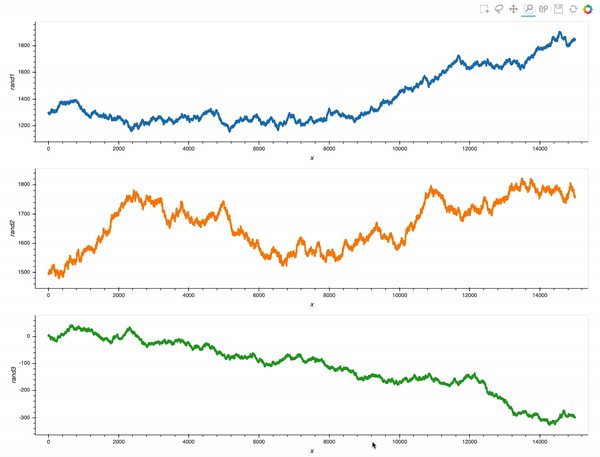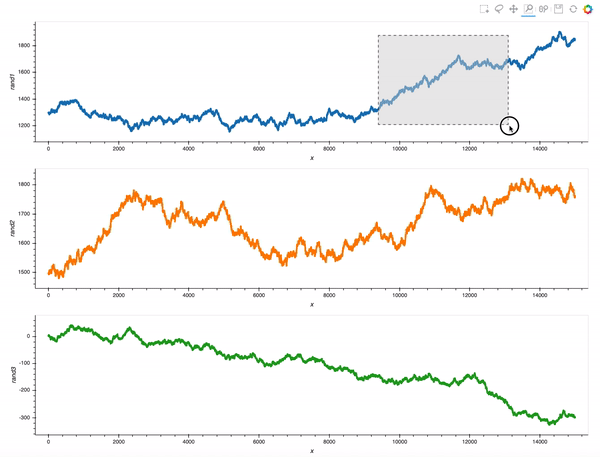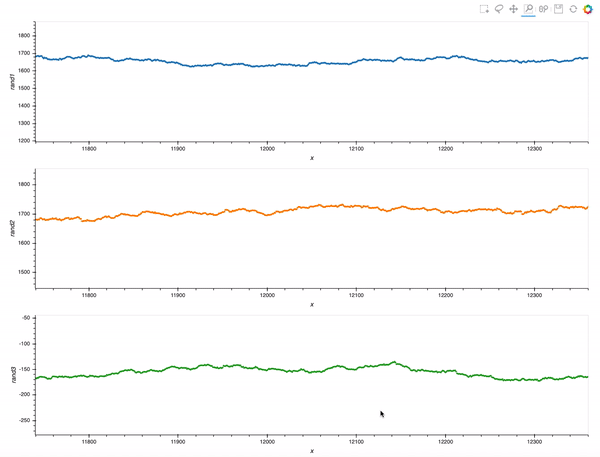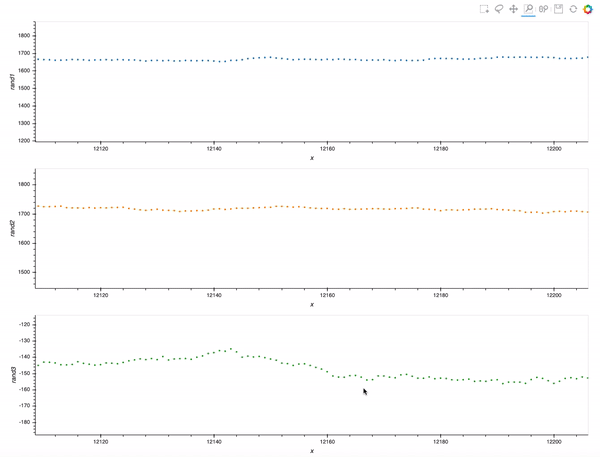
That is already an amazing result for a 20 minutes effort!
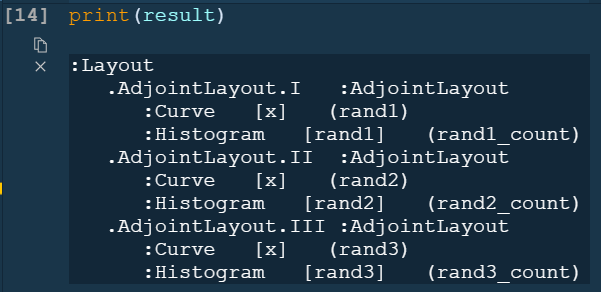
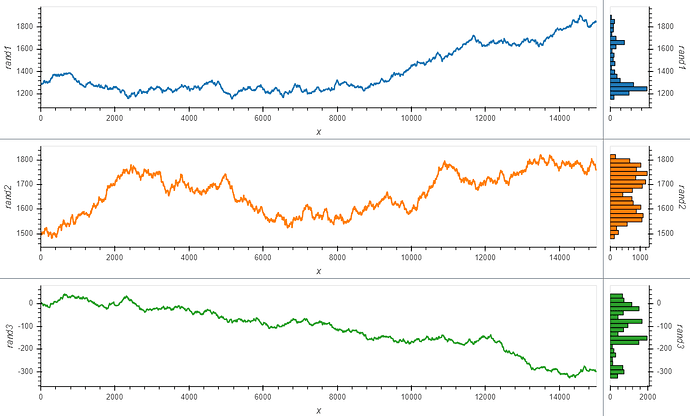
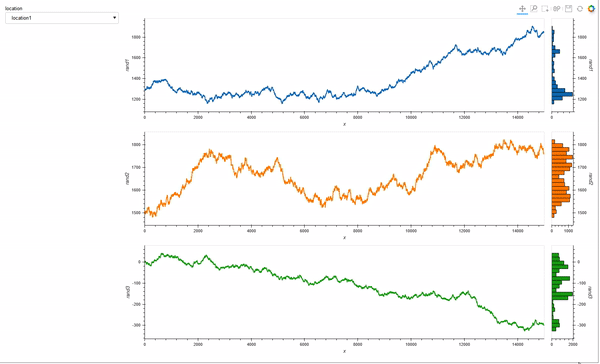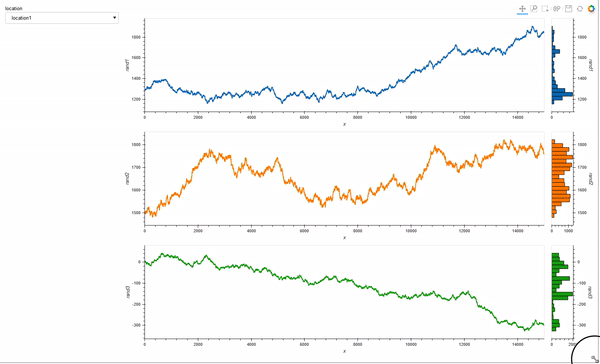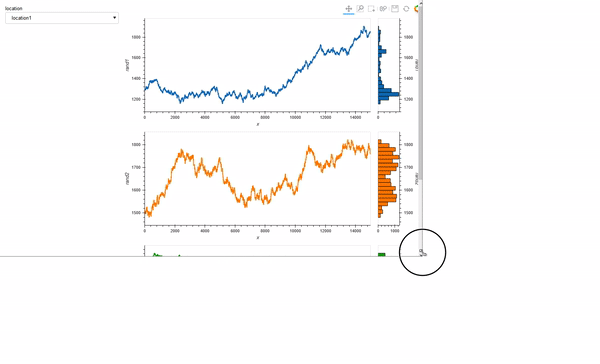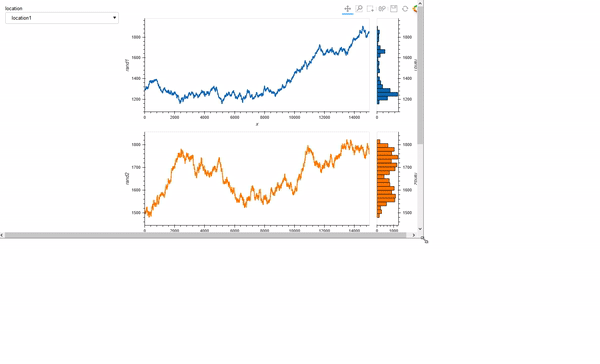
However, what I would really like is to plot a curve rather than points, and also see a histogram, so I adapted the comprehension syntax to work with Curve (after reading the documentation pages Applying customization, and Composing elements):
colors = hv.Cycle('Category10').values
dims = ['rand1', 'rand2', 'rand3']
layout = hv.Layout([hv.Curve(data_df, 'x', dim).opts(height=300, width=1200, color=c).hist(dim) for c,
dim in zip(colors,[d for d in dims])])
link_selections(layout).cols(1)
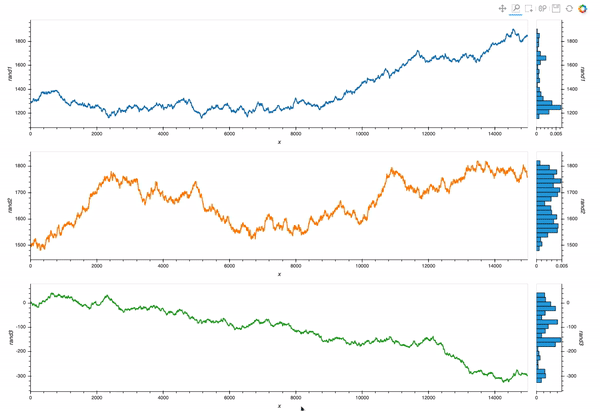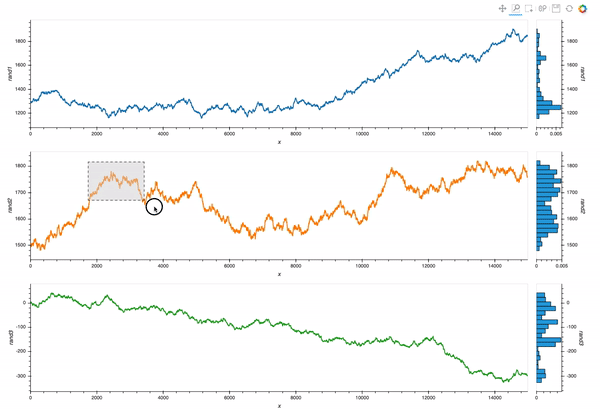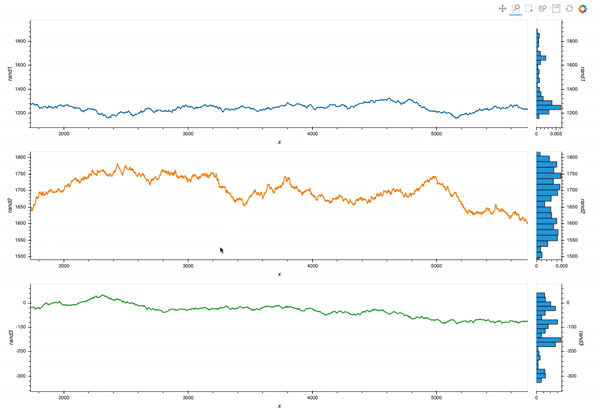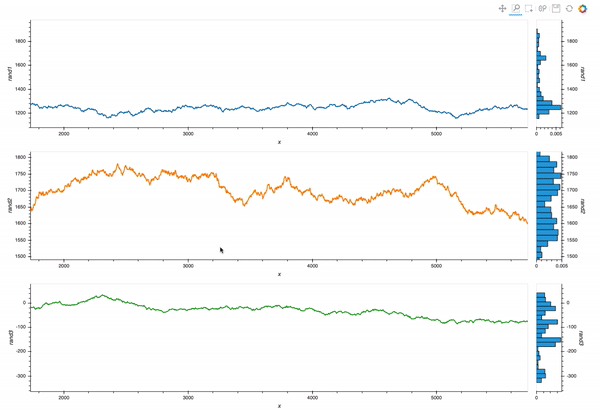
Which is almost exactly what I want. But I still struggle with the different layers of opts syntax.
Question 1: with the comprehension from the last code block, how would I make the histogram share color with the curves?
Now, suppose I want to rasterize the plots (although I do not think is quite yet necessary with 15,000 points like in this case), I tried to adapt my first example with Points:
cmaps = ['Blues', 'Greens', 'Reds']
dims = ['rand1', 'rand2', 'rand3']
layout = hv.Layout([
shade(rasterize(hv.Points(data_df, dims),
cmap=c)).opts(width=1200, height = 400).hist(dims[1])
for c, dims in zip(cmaps, [['x', d] for d in dims])
])
link_selections(layout).cols(1)
This is a decent start, but again I struggle with the options/customization.
Question 2: in the above cod block, how would I pass the colormaps (it does not work as it is now), and how do I make the histogram reflect data values as in the previous case (and also have the right colormap)?
Thank you!