Thank you very much for your kind offer!
The following is a more complete example of how my data looks:
import hvplot.pandas
import numpy as np
import pandas as pd
import xarray as xr
# data
ta = xr.DataArray(
np.random.random((4, 2, 3)),
dims=("time", "lat", "lon"),
coords={"time": np.arange(0, 4), "lat": np.arange(0, 2), "lon": np.arange(0, 3)},
)
tsurf = xr.DataArray(np.random.random((4, 2, 3)), dims=("time", "lat", "lon"))
ptype = xr.DataArray(
np.array([3, 2, np.nan, 1, np.nan, np.nan]).reshape((2, 3)), dims=("lat", "lon")
)
vtype = xr.DataArray(
np.array([np.nan, np.nan, 5, np.nan, 2, 7]).reshape((2, 3)), dims=("lat", "lon")
)
ds = xr.Dataset({"ta": ta, "tsurf": tsurf, "ptype": ptype, "vtype": vtype})
resulting in a Dataset
<xarray.Dataset>
Dimensions: (time: 4, lat: 2, lon: 3)
Coordinates:
* time (time) int64 0 1 2 3
* lat (lat) int64 0 1
* lon (lon) int64 0 1 2
Data variables:
ta (time, lat, lon) float64 0.7546 0.5793 0.1435 ... 0.5606 0.545
tsurf (time, lat, lon) float64 0.164 0.9778 0.8544 ... 0.4033 0.3633
ptype (lat, lon) float64 3.0 2.0 nan 1.0 nan nan
vtype (lat, lon) float64 nan nan 5.0 nan 2.0 7.0
The idea is that I have “maps” (lat and lon) of air temperatures ta above certain surfaces with temperatures tsurf for different times. For each surface (lat and lon), I know its pavement type (ptype) and vegetation type (vtype). Both, ptype and vtype, do not overlap, i.e. if one is not nan, the other must be nan. In my real data, all dimensions are (potentially much) larger and I also have more type-like arrays, i.e. more arrays that are independent of time.
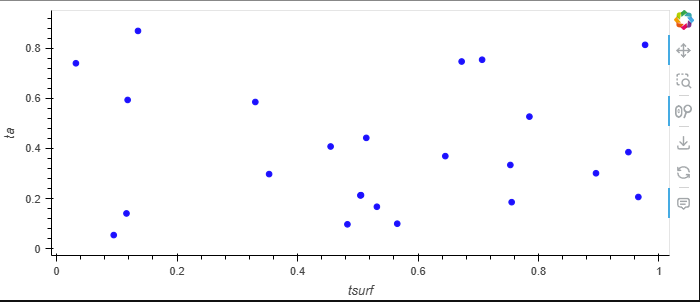
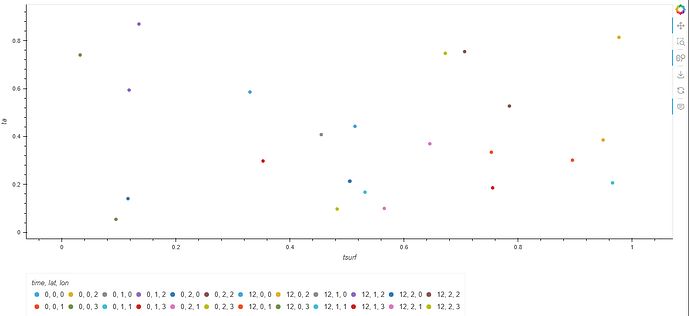
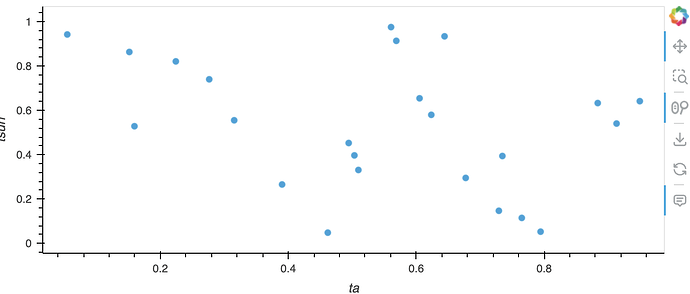
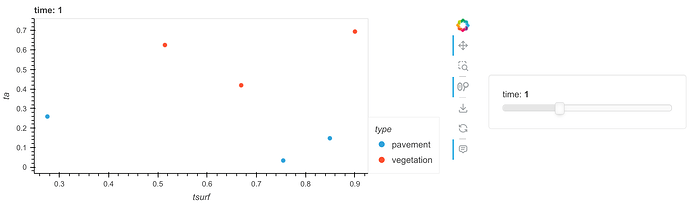
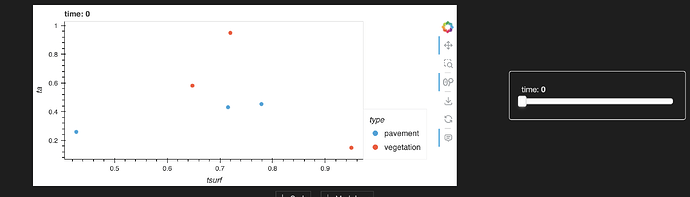
My aim is now to create a scatterplot of ta vs tsurf, by type and each time. I do have a solution for that but it is based on a pandas dataframe:
df = ds.to_dataframe()
# generalization of types
df["type"] = df["ptype"].notna() * 1 + df["vtype"].notna() * 2
df["type"] = (
df["type"].astype("category").cat.rename_categories(["pavement", "vegetation"])
)
df.hvplot.scatter(x="tsurf", y="ta", by="type", groupby="time")
resulting in
The advantage of pandas is that I can use a categorical dtype for type which is not available in xarray (please correct me if I am wrong here). The disadvantage is, however, that in order to fill the table, all non-time dependent quantities are replicated resulting in a unnecessarily large dataset:
ta tsurf ptype vtype type
time lat lon
0 0 0 0.754638 0.163989 3.0 NaN pavement
1 0.579287 0.977793 2.0 NaN pavement
2 0.143468 0.854425 NaN 5.0 vegetation
1 0 0.001652 0.898264 1.0 NaN pavement
1 0.746001 0.787206 NaN 2.0 vegetation
2 0.305514 0.316464 NaN 7.0 vegetation
1 0 0 0.327031 0.647004 3.0 NaN pavement
1 0.458336 0.968298 2.0 NaN pavement
2 0.892447 0.711597 NaN 5.0 vegetation
1 0 0.839581 0.674225 1.0 NaN pavement
1 0.914335 0.250300 NaN 2.0 vegetation
2 0.592095 0.444696 NaN 7.0 vegetation
2 0 0 0.993893 0.067323 3.0 NaN pavement
1 0.551221 0.554406 2.0 NaN pavement
2 0.645342 0.683610 NaN 5.0 vegetation
1 0 0.973098 0.933462 1.0 NaN pavement
1 0.704639 0.053339 NaN 2.0 vegetation
2 0.976332 0.161400 NaN 7.0 vegetation
3 0 0 0.530574 0.550358 3.0 NaN pavement
1 0.139996 0.707700 2.0 NaN pavement
2 0.964032 0.175724 NaN 5.0 vegetation
1 0 0.225253 0.981884 1.0 NaN pavement
1 0.560618 0.403304 NaN 2.0 vegetation
2 0.544982 0.363305 NaN 7.0 vegetation
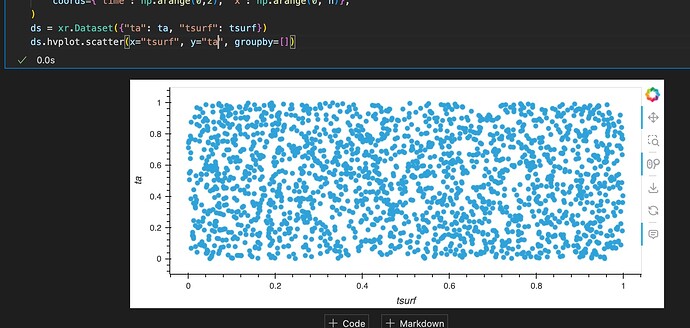
Thus, my idea was to stick with the xarray object. Any idea how to do that? Or any other advice?
![]()